In QGene, simulation of a QTL-mapping population must be preceded by simulation of a map. To simulate trait data, you must first simulate a population with marker data, since trait values are controlled by QTLs and these are modeled as marker loci. While the generated QTLs are invisible to analysis methods, they are still shown on the screen.
Trait simulation, like map and population simulation, is controlled by parameter settings including the value of the random seed.
- Trait name is the name of the trait you want to create. A trait name must start with a letter and should not contain whitespace; instead of spaces, use underscores (_).
- Trait type indicates the data type of your trait. At present QGene simulates only metric traits
- Heritability indicates the proportion of trait variation accounted for by genetic variation.
- Population mean is the overall mean of the trait, taken over all possible genotypes. Don't expect this to match the mean of the population that's actually generated, since depending on the mating design the frequencies of these genotypes may not be balanced. What it refers to is the intercept after genetic effects have been subtracted.
- Noise distribution is the distribution model of the nongenetic influences on the trait. You will usually choose Normal unless you have reason to think that normal noise is not adequate.
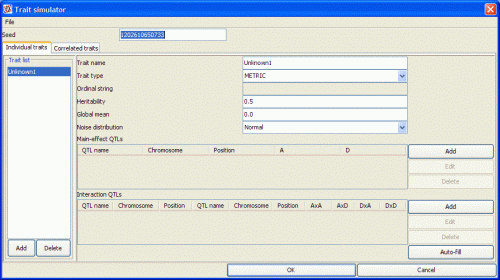
There are two ways to enter the QTLs: manually or automatically. If you let QGene fill in the QTLs automatically, you'll still be able to edit the names, positions, and effects of the automatically generated QTLs as described below. If you prefer manual entry, skip to the subsection below.
Filling in QTLs automatically
- If you want QGene to provide you with an initial set of QTLs, click the Auto-fill button. You'll be presented with the dialog below.
- Number of traits indicates how many traits you want QGene to generate.
- Trait name is the prefix of the trait name. So if you want to generate 3 traits and your trait prefix is "QT", the resulting traits will be named "QT1", "QT2", and "QT3". If there is already a trait called "QT1", your trait names will be "QT2", "QT3", and "QT4". Trait prefixes must also follow the naming rules above.
- Trait type, heritability, global mean or population mean, and noise distribution are the same as above.
- Number of main-effect QTLs and Number of interaction QTLs refer to the number of QTLs of this kind per trait.
- Largest main-effect value and largest interaction-effect value are self-explanatory.
- Effect behavior tells QGene how to generate the effects.
- If it's "FIXED", the values you mentioned above are the effect values for all QTLs generated.
- If it's "RANDOM", QGene will randomize the effect value from half of the value to the value specified. For example, if you specify the largest main effect value to be 10 and you choose random effect behavior, QGene will choose at random a number between 5 and 10. This also applies to interaction effects.
- If it's "DECAY", the first QTL QGene generated will have the effect value specified. The second and subsequent QTLs will have progressively diminishing effect values.
- Make traits correlated (currently disabled) is a feature to create correlated traits. It can be turned on only if you want to generate more than one trait. By checking this option, you can specify how many QTLs are jointly responsible for all traits you are about to generate.
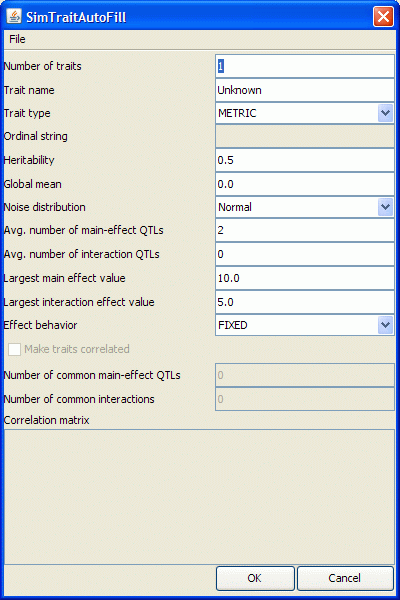
If you click OK on Auto-fill dialog, the table below Main-effect QTLs and Interaction QTLs will be filled with some entries, depending on your specification. Now you can do manual adjustments on the auto-generated QTLs. If you're happy with the chosen parameters, you may skip the editing step.
Manual QTL input/editing
- You can click the Add button to add main-effect or interaction QTLs and edit their attributes yourself. The Edit button lets you edit the attributes of the highlighted QTL. The Delete button lets you remove selected QTLs.
- If you add or edit a QTL, another dialog will appear. You may change the QTL name, the chromosome where it resides, its position in the chromosome (in centiMorgans), and its effect values.
- You may want to review all attributes of the traits you want to generate. In the first image above, you'll see a Trait list panel on the left. You may click on the entries to see the attributes of the traits.
- Once you're satisfied, click OK.
How to select values for QTL effects
If you don't know what size of effect to use, you should try the QTL effects calculated from analysis of a real data set. For example if you are simulating seed size you might use the additive effects from a QTL based on CIM or SIM analysis, recalling that these effects are expressed in the same unit as the trait (for example, mm).
How QGene computes trait values
Let's suppose you want to generate a trait with two QTLs (A and B) on an F2 population. Suppose there is no interaction between the QTLs and the QTL parameters are as follows:
| QTL | Additive effect | Dominant effect |
|---|---|---|
| A | 10 | 3 |
| B | 5 | 0 |
This means that the AA genotype will contribute 10 - 1/2 x 3 = 8.5 to the trait, Aa will contribute +3, and aa will contribute -10 - 1/2 x 3 = -11.5. Likewise, the BB genotype will contribute 5, Bb will contribute 0, and bb will contribute -5.
Suppose we have a population mean of 100. Thus, before QGene adds noise, the trait values are as follows:
| Genotype | Effects | Pop. mean | Trait value |
|---|---|---|---|
| AABB | 8.5 + 5 = 13.5 | + 100 | =113.5 |
| AABb | 8.5 + 0 = 8.5 | + 100 | =108.5 |
| AAbb | 8.5 - 5 = 3.5 | + 100 | =103.5 |
| AaBB | 3 + 5 = 8 | + 100 | =108 |
| AaBb | 3 + 0 = 3 | + 100 | =108 |
| Aabb | 3 - 5 = -2 | + 100 | =98 |
| aaBB | -11.5 + 5 = -6.5 | + 100 | =93.5 |
| aaBb | -11.5 + 0 = -11.5 | + 100 | =88.5 |
| aabb | -11.5 - 5 = -16.5 | + 100 | =83.5 |
Then QGene adds noise according to the distribution and proportional to the heritability parameter.
Interaction effects are computed similarly.