From the main QGene menu, choose File/New map. You'll be presented
with the dialog at right. Most of the options are self-explanatory, so we
provide notes here on only three.
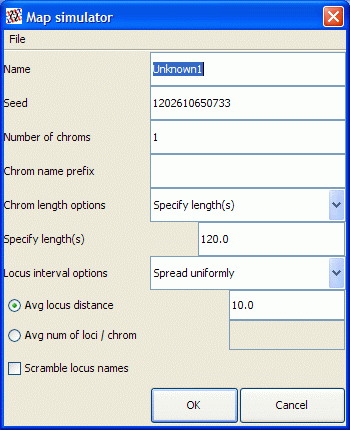
- For chromosome lengths, you can either choose Specify length(s) and provide a cM number to be applied to every chromosome, or choose Use distribution and allow QGene to draw chromosome lengths from the selected statistical distribution with the specified parameters. If you are unsure of the effects of these parameter choices, you may use QGene's default values or experiment for yourself.
- For the Locus interval options, again you may choose to have
QGene space marker loci according to a distribution on interlocus intervals.
- For Fixed option and if you specify average locus distance, then QGene will space the loci as specified. If you specify average number of loci/chrom, QGene will allocate the markers randomly to the chromosomes.
- For Specify option, you may specify locus positions separated by commas. Keep in mind that you should not exceed the chromosome length.
- For Spread uniformly option, QGene will calculate the expected number of loci that will be generated and spread them evenly over all chromosomes.
- Scramble locus names is an option you may choose in order to have QGene name marker loci without a chromosome prefix and with shuffled locus numbers. The reason you might want to do this is for creating a data set for a student linkage-mapping exercise without making it obvious to which linkage groups the markers belong.